As we enter a new era of the post-industrial revolution characterized by rapid robotization and technological innovation, new horizons are opening that necessitate fresh, interdisciplinary approaches to visioneering. The fusion of quantum physics and biology represents one such pioneering approach, suggesting potential applications that could redefine the limits of both fields.
Exploring the Biofield: A Convergence of Quantum Physics and Biology
The concept of the "Biofield," inspired by the quantum mechanics underlying the Higgs field, proposes an innovative framework for expanding the capacity of genetic information storage within DNA. This speculative idea draws from the principles of quantum field theory, postulating that it is possible to manipulate genetic material to store information exponentially, perhaps even infinitely. This theoretical mechanism envisions inducing complex, stable states within genetic structures akin to the multiple state superpositions observed in quantum particles.
Practical Applications and Implications:
Medicine and Healthcare:
- The realm of personalized medicine could experience a paradigm shift, where treatments are not only tailored to individual genetic profiles but are also dynamically adjustable based on real-time genetic data.
- Advances in gene therapy could include programmable, reversible genetic modifications, offering new treatments for complex genetic disorders with high precision and adaptability.
Agriculture and Environmental Management:
- Genetically modified crops or livestock could possess dynamically adjustable traits, capable of responding instantaneously to environmental changes, thus enhancing sustainability and productivity.
- Enhanced genetic storage capabilities could also revolutionize environmental management, particularly in carbon capture and biofuel production. Organisms might be engineered to optimize their biological processes based on fluctuating environmental inputs, leading to more efficient and adaptable systems.
As we stand on the brink of these outstanding horizons, the integration of concepts from quantum physics into biological systems illustrates the potential for innovative solutions that break traditional disciplinary boundaries. This approach not only promises to extend our technological capabilities but also invites a reevaluation of the methodologies we employ in scientific research and application. The success of such interdisciplinary ventures could fundamentally alter our approach to genetic data storage, medical treatments, and environmental management, marking a significant milestone in the ongoing technological evolution.
DNA
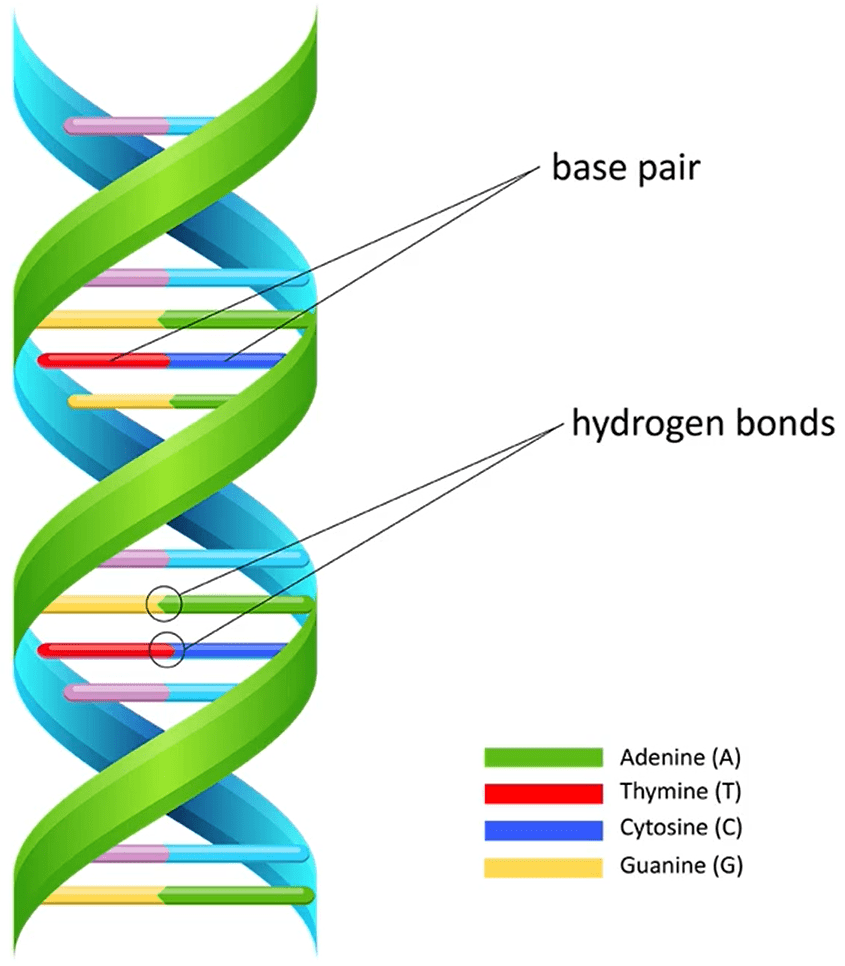
Adenine, guanine, thymine, and cytosine, collectively known as the nucleobases, have several key functions in DNA and RNA. Here's a breakdown of their roles:
- Genetic Information Storage: The primary function of these bases is to store genetic information in cells. The sequence of these bases encodes the instructions for building proteins, which perform most of the functions in living organisms.
- Base Pairing:
- Adenine (A) pairs with thymine (T) in DNA or with uracil (U) in RNA through hydrogen bonds. This pairing is crucial for the double helix structure of DNA and also for the copying of DNA during replication.
- Guanine (G) pairs with cytosine (C) through three hydrogen bonds, which is more than the two bonds between adenine and thymine. This stronger bonding helps stabilize the DNA structure.
Protein Synthesis:
- mRNA Transcription: In the process of transcription, the sequence of bases in DNA is copied to a messenger RNA (mRNA) molecule. Here, thymine (T) in DNA is replaced by uracil (U) in RNA.
- Translation: During translation, the sequence of bases in mRNA is used to determine the sequence of amino acids in proteins. Each set of three bases, called a codon, specifies a particular amino acid.
Regulation of Gene Expression:
- The sequence and arrangement of these bases can also regulate when, how, and how much of a protein is made. This can involve mechanisms such as promoter regions, enhancers, and silencers, which can be located upstream or within a gene sequence.
Genetic Variability and Evolution:
- Mutations, which are changes in the base sequence, can lead to changes in the proteins that are produced. This can affect an organism's phenotype and, consequently, its fitness within an environment. Mutations are a driving force behind evolution.
In summary, adenine, guanine, thymine, and cytosine serve as the foundational elements of genetic code, with their specific pairing ensuring accurate DNA replication and transcription, leading to protein synthesis. They also play crucial roles in regulating gene expression and contributing to genetic diversity and evolution.
The information storage capacity of a genome can be understood by considering the length of the DNA and the number of possible base pair combinations it contains. The total amount of information a genome can store depends largely on the number of base pairs it comprises, and this varies widely among different organisms.
Here's how to conceptualize it in terms of data storage:
- Base Pair as a Unit of Information: Each base pair (A-T or G-C) in the DNA sequence can be thought of as a unit of information. Because there are four bases (adenine, cytosine, guanine, thymine), each base pair can be represented by two bits of information (since 2 bits can encode up to four states).
- Calculating Information Storage:
- For example, the human genome contains approximately 3 billion base pairs. Since each base pair can represent 2 bits of information, the total amount of information in the human genome can be calculated as
- 3 billion base pairs×2 bits per base pair
- 3 billion base pairs×2 bits per base pair.
- This calculation gives 6 billion bits, which converts to about 750 megabytes of data.
Here is a segment of the genomic sequence from the bacterium Caulobacter crescentus that I accessed. Interestingly, this bacterium utilizes codons, which are also found in human DNA sequences. This suggests that our DNA might merely be a gateway to a more extensive information storage system existing at a deeper level of reality:
I have created a list of "words" with all the codons and further normalized them:
Now creating a dataframe withe the words to further vizualize the RNA clusters.
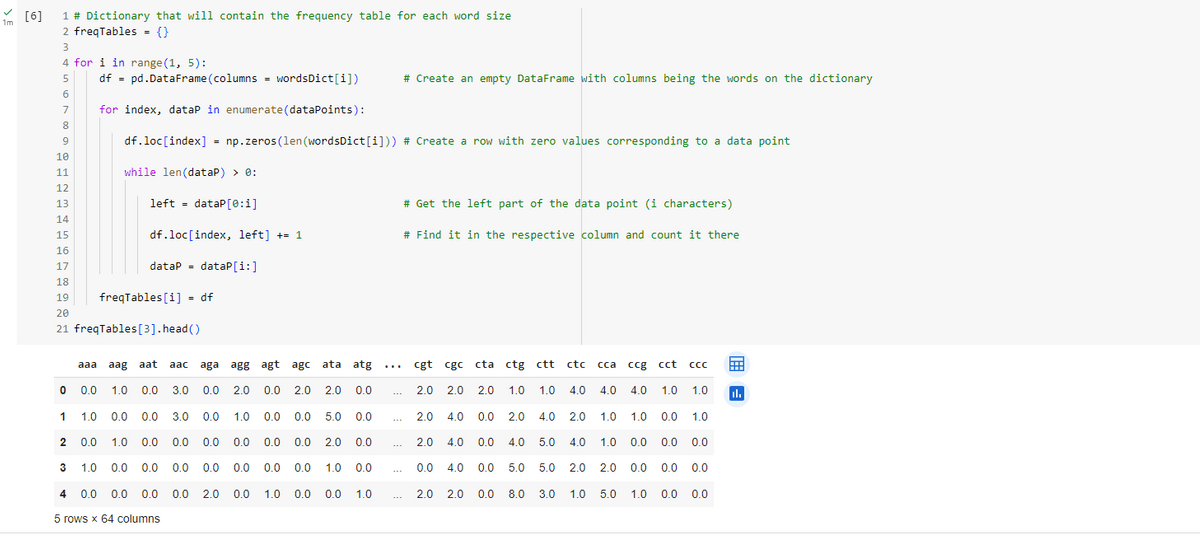
After the PCA analysis we are coming to plotting:
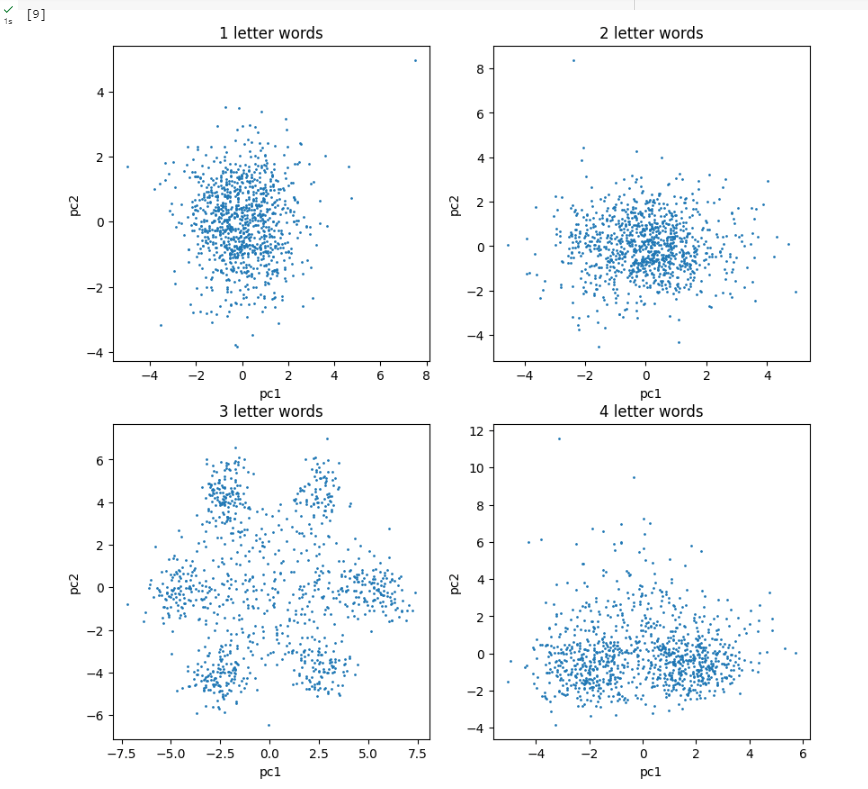
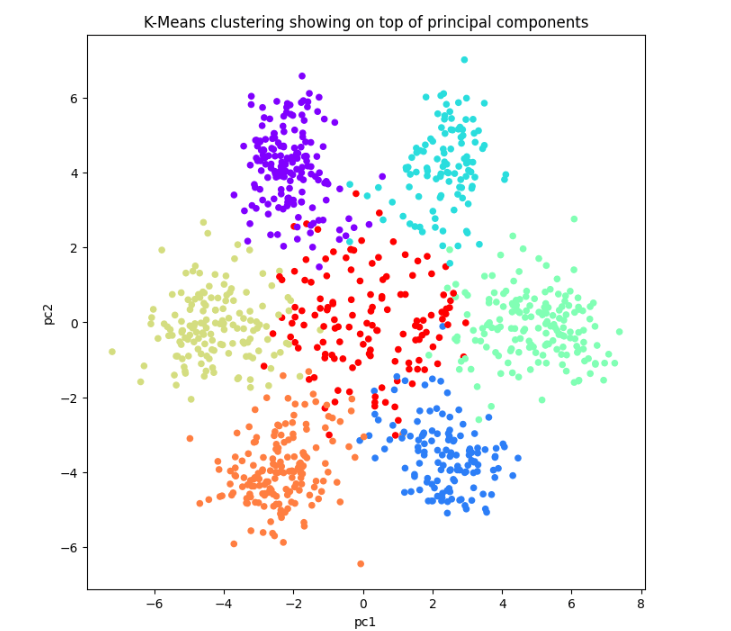
We have used the K-means Clustering unsupervised algorithm to cluster the three-letter gene sequences. Based on our analysis from the previous section, we identify the possibility of 6 or 7 clusters. Given that some genes may not contain information, those that are significantly distant from the six identified centroids might represent non-informative genes. Consequently, we propose that there are 7 clusters, although this hypothesis could be verified by comparing clustering performance using both 6 and 7 clusters.
Consequently, the application of statistical unsupervised learning techniques, specifically Clustering (K-means Clustering) and Dimensionality Reduction (PCA), has enabled us to visualize, confirm, and substantiate the biological finding that the DNA genetic sequence of bacteria is structured into three-letter words known as codons, which intriguingly utilize the same set as humans.
The Higgs field
Further exploring unconventional concepts: As we continue to reflect on the loss of Peter Higgs on April 08, 2024, I ventured into his area of expertise, examining the Higgs field and particularly the Higgs bosons, to explore their application to the biofield concept we are investigating.

The "Mexican hat" or "wine bottle" shape of this potential allows the field to acquire a non-zero vacuum expectation value (VEV). When the Higgs field settles into this minimum energy configuration, it breaks the electroweak symmetry of the Standard Model in a process known as spontaneous symmetry breaking.
Particle Mass Acquisition
The Higgs field interacts with other particles, such as the W and Z bosons and fermions (like electrons and quarks), through Yukawa couplings. The strength of these interactions determines the mass of these particles. The Lagrangian that describes these interactions and mass acquisition can be expressed as:

Gauge Boson Masses
The Higgs field gives mass to the W and Z bosons through its interactions. Before symmetry breaking, these bosons are massless. However, when the Higgs field acquires a VEV, these bosons interact with the non-zero components of the field, gaining mass proportional to the strength of their coupling to the Higgs field. This is described by:
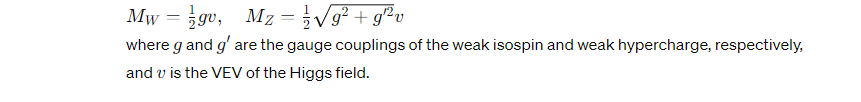
Quantum Fluctuations and the Higgs Boson
The physical Higgs boson arises as a quantum excitation of the Higgs field around its vacuum expectation value. This can be seen as fluctuations around the VEV in the potential landscape. The mass of the Higgs boson itself is derived from the curvature of the potential at the VEV, given by:
These elements together illustrate how the Higgs mechanism provides a coherent mathematical explanation for the origins of mass in elementary particles. The discovery of the Higgs boson at CERN in 2012 provided experimental confirmation of this theory, a cornerstone of particle physics.
Now let us apply the analogy of the Higgs field to the biofield arrays focusing specifically on the theoretical analogy with the Higgs boson, exploring how smaller biological particles similar to the Higgs boson might affect DNA and molecular biology.
Theoretical Framework of the "Biofield" Potential
Just like the Higgs field, let's consider a new kind of field in molecular biology, which we'll call the "Biofield." This field could interact with the components of DNA, potentially nucleobases, influencing genetic expression and stability. The Biofield would have a potential similar to the Higgs potential, described by a "Mexican hat" shape, which allows for the breaking of symmetries and the emergence of new properties.
Biofield Potential Formula:

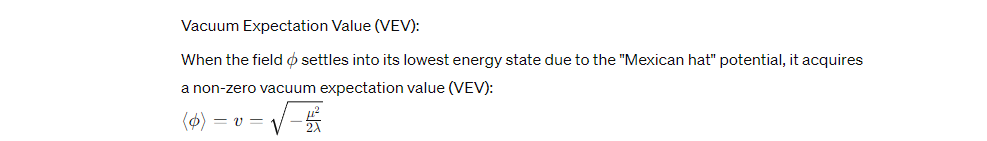
This VEV is crucial as it represents the field's stable state, around which the particles (possibly components of DNA or RNA) gain novel properties or interactions.
Interaction with Biological Fermions (e.g., Nucleobases)
We introduce a Yukawa-like interaction between the Biofield and biological components, such as nucleobases in DNA. This interaction could alter the properties of these biological molecules, potentially affecting their role in genetic processes.
Interaction Lagrangian:

Full Theoretical Model:
Combining these elements, the full Lagrangian for the system including the Biofield and its interactions with DNA components would be:
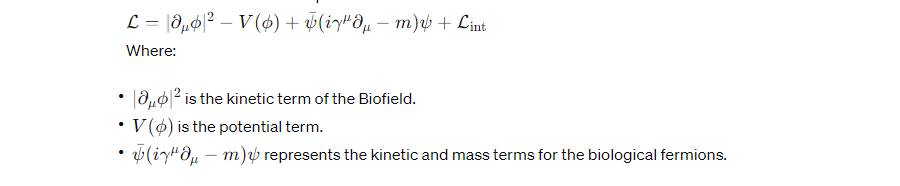
Implications of this Model:
- Enhanced Genetic Information Storage: If such interactions allow DNA to store more information within the same physical space, or even utilize information in new ways, this could vastly increase the complexity and capability of genetic systems.
- Novel Genetic Mechanisms: New mechanisms for gene regulation, expression, and mutation correction could emerge, similar to discovering a new layer of genetic control.
- Advanced Biotechnological Tools: More precise genetic manipulation techniques could be developed, allowing for targeted therapies and advanced genetic engineering capabilities.
This theoretical exploration, drawing an analogy with the Higgs boson and its mechanism, provides a speculative yet fascinating viewpoint on how fundamental new discoveries in molecular biology might emerge. It's important to note, however, that this is a highly theoretical approach and actual biological systems may behave differently due to their complex and varied nature.
The theoretical application of a Higgs-like field mechanism—termed here as the "Biofield"—to genome information storage presents an intriguing frontier in synthetic biology and biophysics. While this concept is speculative and primarily exploratory, it potentially opens up a new paradigm in how information is stored and manipulated within biological systems. By postulating that interactions within the Biofield could modulate genetic expression and stability, we envision a system where the complexity and capacity of genetic information storage are not merely enhanced but expanded exponentially, potentially to infinity.
Potential Infinite Expansion of Genetic Storage
In theory, if the Biofield can induce varying states or configurations in genetic material (akin to multiple stable states in quantum systems), each configuration could represent different levels of information. Unlike traditional binary systems which are limited to two states (0 and 1), this system could leverage a vast array of states, exponentially increasing the storage capacity. This could be likened to quantum computing where qubits hold a superposition of multiple states, offering a parallel in biological systems.
Practical Applications
1. Medicine and Healthcare:
- Personalized Medicine: With vastly expanded genetic storage capabilities, more complex genetic information could be stored about an individual, leading to highly personalized medical treatments. Information about individual responses to different treatments could be encoded directly within one's genome, accessible and modifiable in real-time as treatment responses are observed.
- Gene Therapy: Enhanced storage could revolutionize gene therapy, allowing for more complex genetic modifications which are reversible or adjustable over time. This could be crucial for treating genetic diseases where multiple genes are involved or where gene expression needs to be tightly controlled.
2. Biology and Agricultural Science:
- Synthetic Biology: Biofield applications could lead to the development of new biological systems or organisms with entirely novel properties, programmed and managed through enhanced genetic storage systems.
- Agricultural Improvements: Genetically modified crops or livestock with dynamically adjustable traits (e.g., resistance to environmental stresses) could be developed, where the genetic traits are modulated based on changing environmental conditions.
3. Transportation:
- Biofuel Production: Genetically engineered organisms with expanded genetic storage capabilities could be designed to produce biofuels more efficiently. These organisms could dynamically adjust their metabolic pathways based on feedstock availability or environmental conditions.
- Environmental Sensing: Vehicles, especially unmanned aerial or ground vehicles, could be equipped with bio-sensors that have dynamically adjustable sensitivity and specificity, based on genetically stored and adjustable parameters.
4. Energy Sector:
- Biological Energy Storage: Microorganisms or plants could be engineered to store energy in new ways or to increase their natural energy storage capacities, leveraging their genetic material as a dynamic system that adjusts to energy demands and storage conditions.
- Carbon Capture and Conversion: Organisms could be engineered to not only capture carbon dioxide more efficiently but also to store information on conversion processes genetically. This might allow them to switch between different methods of carbon fixation and conversion based on environmental inputs.
Conclusion
The application of a Biofield to expand genome information storage effectively to infinite capacity presents a transformative but speculative leap in technology that bridges particle physics, biology, and information science. While the practical implementation of such a system remains a formidable challenge, the theoretical potential it offers could revolutionize multiple fields, providing tools that are more adaptable, efficient, and capable than currently imaginable.
This exploration, although grounded in theoretical physics, encourages a multidisciplinary approach to research and development in genetic science, suggesting that the principles underlying the universe's most fundamental particles might find unexpected applications in the very fabric of biological life. As with all speculative technologies, rigorous scientific validation and ethical considerations must guide further exploration and potential development.


